Teaching experience and interests
My teaching interests are in the broad field of Statistical Physics, condensed-matter physics and biophysics. At the university of Orléans I am currently giving the following courses:- Introduction to condensed matter physics
- Introduction to elasticity theory
- Selected topics in theoretical biophysics
- Mathematical methods for the physical sciences
Teaching material
Selected topics on protein dynamics
Lectures delivered in November 2013 at Tsinghua University, Beijing and in November 2016 at UACM, Mexico city
Selected topics in diffusion
Lectures delivered at the University of Florence, ERASMUS Ph.D. course, 2012 and 2013
Statistical analysis of DNA sequences
Lecture delivered in July 2002 at: International IP-SOCRATES summer school on nonlinear time series analysis
Selected topics in physical biology, Ph.D. course (Tsinghua University, Beijing, UACM, Mexico city and University of Orléans)
This course is intended to present some recent advances in physical biology, covering mainly two topics, (i) coarse-grained models of protein dynamics and (ii) diffusion- limited bimolecular reactions with an emphasis on di usion in crowded and con ned environments. The course is open to all Ph.D. students in the scientifc doctoral schools. A background in elementary statistical mechanics and calculus is required but no pre- requisite in molecular and cell biology is needed, as a basic introduction to this will be given. The most complex mathematical derivations are done in full length and at slow pace, covering all details in a self-contained manner. The course is structured in about 8 courses of 2 hours, two of which can be organized as exercise classes, with the possibility of both supervised problem solving and running individual or group sessions of computer simulations and calculations, covering aspects treated during the lectures. The material for these (computer codes etc.) are supplied by the instructor. Students have the possibility to set up their personal laptops to use them for the exercise classes.
-
 LECTURE 1:
LECTURE 1:
Introduction to the most important biomolecules in molecular and cell biology. A personal account on the basic building blocks of biological matter. -
 LECTURE 2:
LECTURE 2:
Statistical physics models of DNA and basic concepts of polymer physics. The case of DNA pulling by optical tweezers. -
 LECTURES 3-4:
LECTURES 3-4:
- Modeling protein dynamics I. Force fields and normal mode analysis.
- Modeling protein dynamics II. Elastic network models, an account on how exceedingly simple modeling schemes still teach a lot on biologically relevant issues.
-
 LECTURES 5-8
LECTURES 5-8
- Diffusion-limited bimolecular reactions I. Modeling reactions in the liquid phase through boundary value problems. The classic Smoluchowski and Debye theories.
- Diffusion-limited bimolecular reactions II. Cells uniformly covered in receptors versus cells with a single, large cluster of receptors. Which is better? A (not so?) surprisingly simple answer from a complex (but instructive) mathematical model.
-
Diffusion-limited bimolecular reactions III. The concept of
diffusive interaction. Are two closely spaced receptors as effective as two separated ones, more or less effective? - Diffusion-limited bimolecular reactions IV. Towards realistic modeling of transport in the cell. Selected topics in diffusion in crowded environments. What do simple stochastic processes teach us on this subject?
Cours introduction au calcul scientifique, L2 Universite d'Orleans
-
 Script to generate N random numbers distributed uniformly
in the interval [0,1] and N random numbers distributed
exponentially in [0,Infinity] (needs numpy)
Script to generate N random numbers distributed uniformly
in the interval [0,1] and N random numbers distributed
exponentially in [0,Infinity] (needs numpy)
-
 Script to compute the histogram and the cumulative distribution
of a set of data (needs numpy)
Script to compute the histogram and the cumulative distribution
of a set of data (needs numpy)
-
 Script to integrate the logistic differential equation.
Using simple forward Euler scheme (needs numpy)
Script to integrate the logistic differential equation.
Using simple forward Euler scheme (needs numpy)
-
 An example of Monte Carlo simulation with the
Metropolis scheme with integrand f(x) = x*x.
Example from Tao Pang, Introduction to Computational Physics
Cambridge University Press (2006).
An example of Monte Carlo simulation with the
Metropolis scheme with integrand f(x) = x*x.
Example from Tao Pang, Introduction to Computational Physics
Cambridge University Press (2006).
-
 Numerical integration of a function of one variable with
the trapezoidal and midpoint rules. The script plots the error as a function of
the integration step to verify that both algoriyhms are O(h**2).
Numerical integration of a function of one variable with
the trapezoidal and midpoint rules. The script plots the error as a function of
the integration step to verify that both algoriyhms are O(h**2).
-
 Sample vector of random deviates from a uniform distribution
Sample vector of random deviates from a uniform distribution
Sample vector of random deviates from a Gaussian distribution -
 Numerical integration of Lagrange equation for a ball rolling wuthout friction down
a trail whose equation H(x) is given analytically. The script uses Heun algorithm
(a.k.a. predictor-corrector or modified Euler).
Numerical integration of Lagrange equation for a ball rolling wuthout friction down
a trail whose equation H(x) is given analytically. The script uses Heun algorithm
(a.k.a. predictor-corrector or modified Euler).
-
 Numerical integration of a stochastic differential equation with multiplicative noise,
of the form dx/dt = F(x,t) + G(x,t)xi, where xi is a white noise process.
The script uses Heun algorithm
(a.k.a. predictor-corrector or modified Euler).
Numerical integration of a stochastic differential equation with multiplicative noise,
of the form dx/dt = F(x,t) + G(x,t)xi, where xi is a white noise process.
The script uses Heun algorithm
(a.k.a. predictor-corrector or modified Euler).
Cours de langage et calcul scientifique, L3 Universite d'Orleans
-
 Lecture 1
Lecture 1
-
 Lecture 2
Lecture 2
-
 Lecture 3
Lecture 3
-
 Lecture 4
Lecture 4
-
 Lecture 5
Lecture 5
-
 Lecture 1
Lecture 1
-
 Lecture 7
Lecture 7
-
 Lecture 8
Lecture 8
-
 FORTRAN 90 code for molecular dynamics simulations.
FORTRAN 90 code for molecular dynamics simulations.
-
 FORTRAN 90 code for discrete Fourier transform.
FORTRAN 90 code for discrete Fourier transform.
-
 FORTRAN 90 code
to integrate Gompertz's model (simple Euler O(1)).
FORTRAN 90 code
to integrate Gompertz's model (simple Euler O(1)).
-
 FORTRAN 90 code
to integrate Gompertz's model (simple Euler O(1), modular architecture with subroutines
and functions).
FORTRAN 90 code
to integrate Gompertz's model (simple Euler O(1), modular architecture with subroutines
and functions).
-
 FORTRAN 90 code
to integrate Gompertz's model (modified Euler O(2), predictor-corrector).
FORTRAN 90 code
to integrate Gompertz's model (modified Euler O(2), predictor-corrector).
-
 Sample FORTRAN 90 code
to integrate a one-dimensional flow displaying two saddle-point bifurcations (modified Euler O(2))
Sample FORTRAN 90 code
to integrate a one-dimensional flow displaying two saddle-point bifurcations (modified Euler O(2))
-
 FORTRAN 90 code to integrate a one-dimensional flow
using functions and subroutines (modified Euler O(2)). The code also computes the nearest fixed
point to the initial condition (steepest descent) and performs a linear stability analysis around
this point. The code is meant to illustrate the idea of large-amplitude branches of fixed points.
FORTRAN 90 code to integrate a one-dimensional flow
using functions and subroutines (modified Euler O(2)). The code also computes the nearest fixed
point to the initial condition (steepest descent) and performs a linear stability analysis around
this point. The code is meant to illustrate the idea of large-amplitude branches of fixed points.
-
 FORTRAN 90 code to integrate a one-dimensional stochastic flow
with the Milshtein method. This example illustrates the simple (but non-trivial!) linear
stochastic equation with multiplicative noise. This code outputs a file with a single
stochastic trajectory labelled by the seed used.
FORTRAN 90 code to integrate a one-dimensional stochastic flow
with the Milshtein method. This example illustrates the simple (but non-trivial!) linear
stochastic equation with multiplicative noise. This code outputs a file with a single
stochastic trajectory labelled by the seed used.
-
 FORTRAN 90 code to integrate a one-dimensional stochastic flow
with the Milshtein method. This example illustrates the simple (but non-trivial!) linear
stochastic equation with multiplicative noise. This code performs averages over a given
number of independent stochastic trajectories and outputs the average trajectory and its
standard error on the mean.
FORTRAN 90 code to integrate a one-dimensional stochastic flow
with the Milshtein method. This example illustrates the simple (but non-trivial!) linear
stochastic equation with multiplicative noise. This code performs averages over a given
number of independent stochastic trajectories and outputs the average trajectory and its
standard error on the mean. -
 FORTRAN 90 code to integrate a two-dimensional linear
dynamical system with the predictor-corrector (modified Euler) method.
This code is inspired by the famous paper by Steven Strogatz: Love affairs
and differential equations, Math. Magazine, 61, 35 (1988).
FORTRAN 90 code to integrate a two-dimensional linear
dynamical system with the predictor-corrector (modified Euler) method.
This code is inspired by the famous paper by Steven Strogatz: Love affairs
and differential equations, Math. Magazine, 61, 35 (1988). -
 FORTRAN 90 code
to integrate a 2D nonlinear system of the kind
{\dot(x1) = F1(x1,x2),\dot(x2) = F2(x1,x2)}.
The code uses a 4th order Runge-Kutta algorithm.
This example illustrates the case of a non-hyperbolic fixed
point, a center which is changed into a spiral by arbitrarily
small nonlinear terms (proportional to the parameter a, whose
sign controls the stability of the spiral). This example is
example 6.3.2 in S. Strogatz's book "Nonlinear dynamics and chaos"
The code can be easily generalized to higher dimension and to
different nonlinear systems.
FORTRAN 90 code
to integrate a 2D nonlinear system of the kind
{\dot(x1) = F1(x1,x2),\dot(x2) = F2(x1,x2)}.
The code uses a 4th order Runge-Kutta algorithm.
This example illustrates the case of a non-hyperbolic fixed
point, a center which is changed into a spiral by arbitrarily
small nonlinear terms (proportional to the parameter a, whose
sign controls the stability of the spiral). This example is
example 6.3.2 in S. Strogatz's book "Nonlinear dynamics and chaos"
The code can be easily generalized to higher dimension and to
different nonlinear systems.
-
 FORTRAN 77 code to integrate a tabulated function (Simpson)
FORTRAN 77 code to integrate a tabulated function (Simpson)
-
 FORTRAN 90 code to compute the relaxation to
equilibrium of the money distribution in a system of agents trading pairwise according
to microscopic rules preserving the total amount of money in the system. Based on the paper:
FORTRAN 90 code to compute the relaxation to
equilibrium of the money distribution in a system of agents trading pairwise according
to microscopic rules preserving the total amount of money in the system. Based on the paper:
-
A. Dragulesku and V. M. Yakovenko
Statistical mechanics of money
Eur. Phys. J. B - 17 723-729 (2000)
 PDF file
PDF file
-
 LAPACK subroutine DSYEV + dependencies
LAPACK subroutine DSYEV + dependencies
-
NORMAL MODE ANALYSIS
Example: HIV-1 PROTEASE, PDB code: 4HVP
Elastic network model, alpha-C coarse-graining, sharp cutoff interactions (Rc = 10 Ang)
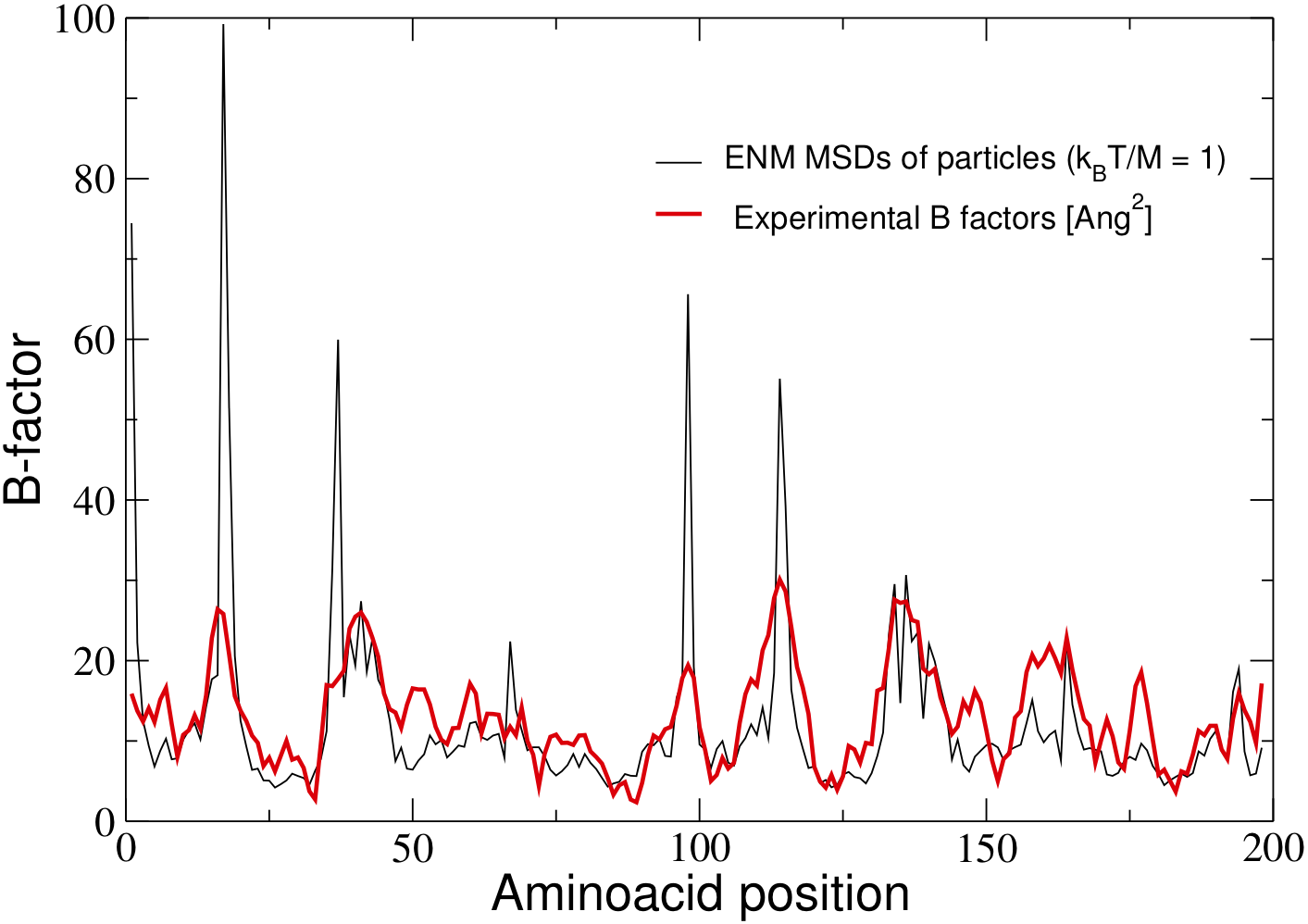
Code
Generic Fortran 90 code, two arguments: PDB file and cutoff [Ang]
Data
NM frequencies (cm^-1)
Normalized displacement pattern of the highest-frequency NM
Normalized displacement pattern of the lowest-frequency NM
Course on Introduction to numerical solution of partial differential equations: the diffusion equation
-
 Python script to solve the 1D diffusion equation in the interval [0,L]
with a simple explicit method. The example illustrates diffusion through
a slab (e.g. the skin stratum corneum) kept at a fixed concentration at its
leftmost side (e.g. the skin/air interface) and in contact with a sink at
its rightmost side (e.g. diffusion into the epidermis).
Python script to solve the 1D diffusion equation in the interval [0,L]
with a simple explicit method. The example illustrates diffusion through
a slab (e.g. the skin stratum corneum) kept at a fixed concentration at its
leftmost side (e.g. the skin/air interface) and in contact with a sink at
its rightmost side (e.g. diffusion into the epidermis).
-
 Python script to solve the 1D diffusion equation in the interval [0,L]
with the Crank-Nicholson method. The example illustrates the time evolution
of a sinusoidal profile, constant Dirichlet boundary conditions at x=0 and x=L.
Python script to solve the 1D diffusion equation in the interval [0,L]
with the Crank-Nicholson method. The example illustrates the time evolution
of a sinusoidal profile, constant Dirichlet boundary conditions at x=0 and x=L.
-
 A short note
describing the methods implemented above.
A short note
describing the methods implemented above.
Some useful Gnuplot scripts
-
 Gnuplot script
to generate a postscript image of a 2D vector field. The components of the field are specified as
functions of (x,y) and the arrows are color-coded according to the local strentgh of the field
(i.e. the norm). The user can also choose to plot the direction field, i.e. the normalized vector
field - same color coding but arrows of the same length at each point.
Gnuplot script
to generate a postscript image of a 2D vector field. The components of the field are specified as
functions of (x,y) and the arrows are color-coded according to the local strentgh of the field
(i.e. the norm). The user can also choose to plot the direction field, i.e. the normalized vector
field - same color coding but arrows of the same length at each point.
-
 Gnuplot script
Basic gnuplot script to generate a cool-looking encapsulated postscript figure for your reports.
Gnuplot script
Basic gnuplot script to generate a cool-looking encapsulated postscript figure for your reports.
Cours de physique statistique avancée, EPFL, a.a. 2004/2005
-
 Equation de langevin pour un oscillateur harmonique
Equation de langevin pour un oscillateur harmonique
-
 Exercise 1: équation de Smoluchowski
Exercise 1: équation de Smoluchowski
-
 Exercise 2: equation de Langevin revisitée
Exercise 2: equation de Langevin revisitée
 Solution
Solution
-
 Exercise 3: processus stochastique simple
Exercise 3: processus stochastique simple
 Solution
Solution
-
 Exercise 4: équation de Fokker-Planck avec pot. harmonique
Exercise 4: équation de Fokker-Planck avec pot. harmonique
 Solution
Solution
-
 Exercise 5: distribution Gaussienne multi-normale
Exercise 5: distribution Gaussienne multi-normale
 Solution
Solution
-
 Exercise 6: rate de transition revisité
Exercise 6: rate de transition revisité
 Solution
Solution
-
 Exercise 7: diffusion sur une membrane (photo-bleaching)
Exercise 7: diffusion sur une membrane (photo-bleaching)
 Solution
Solution
Corso di Fisica generale I, Università di Firenze, 2002-2003
Facoltà di Ingegneria dell'ambiente e delle risorse - PIN Prato (a.a. 2002/2003)-
 Programma del corso
Programma del corso
-
 Elementi di calcolo vettoriale
Elementi di calcolo vettoriale
-
 Elementi di cinematica del punto materiale
Elementi di cinematica del punto materiale
-
 Esercizi di ricapitolazione di cinematica del punto materiale
Esercizi di ricapitolazione di cinematica del punto materiale
-
 Dinamica del punto materiale - Macchine di Atwood e di Fletcher -
Esercizi di ricapitolazione
Dinamica del punto materiale - Macchine di Atwood e di Fletcher -
Esercizi di ricapitolazione
-
 Alcuni esercizi sulla conservazione dell'energia meccanica.
Alcuni esercizi sulla conservazione dell'energia meccanica.
-
Testi dei compiti parziali proposti agli studenti nell'a.a. 2002/2003
 (29/11/2002)
(28/01/2003)
(20/02/2003)
(29/11/2002)
(28/01/2003)
(20/02/2003)